The Rmarkdown source code html for all tutorials is available in the Github indexpage. Helvetica-infected and pathogen-free DNA samples were used for 16S rRNA amplicon sequencing and microbiome analysis.
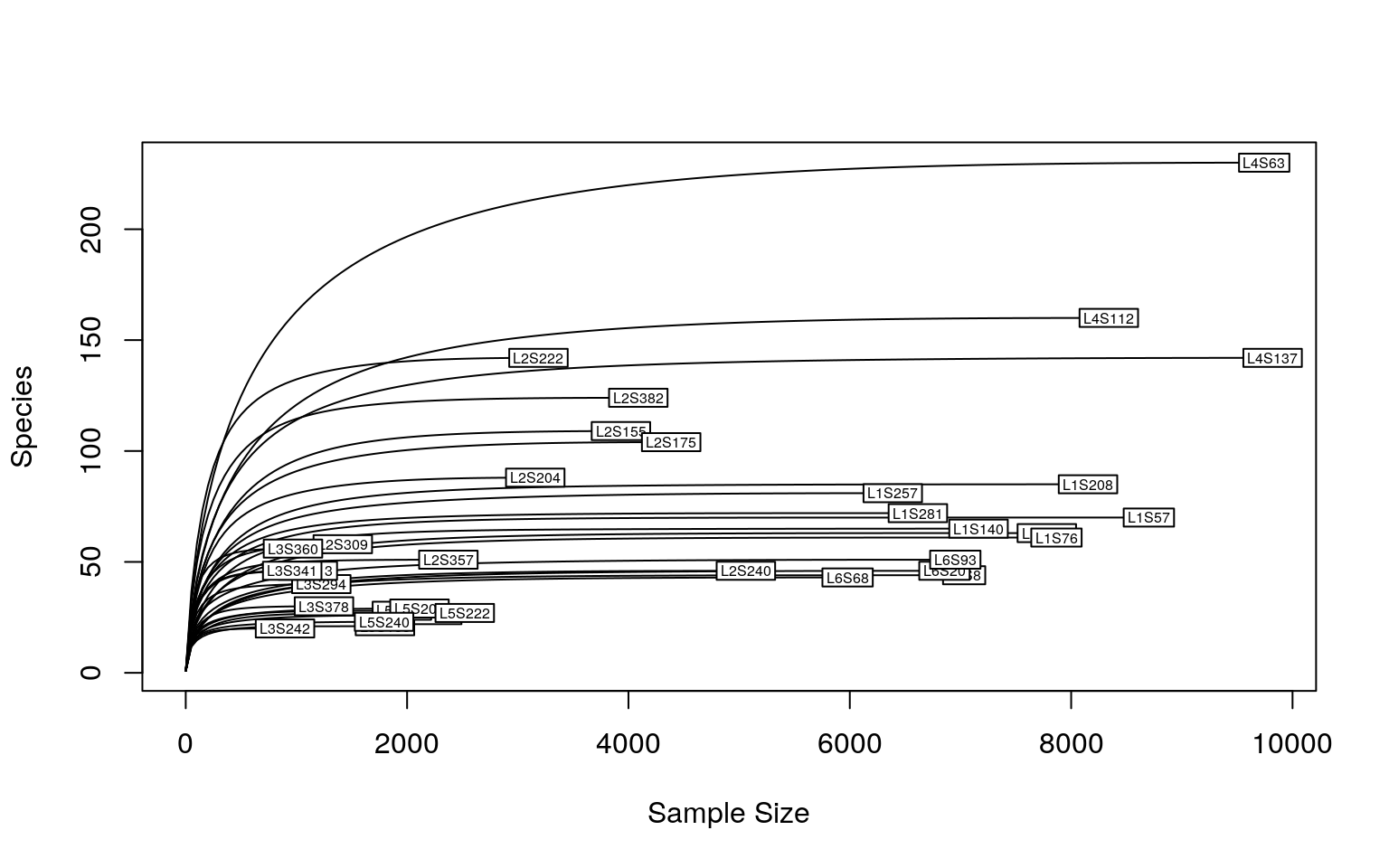
Tutorial For Microbiome Analysis In R Yan Hui
Below you will find R code for extracting alpha diversity beta diversity and taxa abundance.
. However it is still difficult to perform data mining. More specifically the downstream processing of raw reads is the most time consuming and mentally draining stage. 2014 Lahti et al.
Last updated almost 2 years ago. However many of the microorganisms living in complex environments eg. Reading data The first step in any analysis is getting your data into R.
We will do the following in this. This vignette provides a brief overview with example data sets from published microbiome profiling studies Lahti et al. Please follow the below tutorial.
Tools for microbiome analysis in R. Environment initiation Read in your data and select samples for analysis Variable examination and modification Data summary and assessment Taxon prevalence estimations and filtering Data transformation Subsetting Community composition plotting Alpha diversity analysis Beta diversity analysis. Please install the required software and download the example data before coming to the workshop.
Acknowledgements Kindly cite this work as follows. We also provide examples of supervised analyses using random forests and nonparametric testing using community networks and the ggnetwork package. Here we describe in detail and step by step the process of building analyzing and visualizing microbiome networks from operational taxonomic unit OTU tables in R and RStudio using several different approaches and extensively commented code snippets.
High-throughput sequencing of PCR-amplified taxonomic markers like the 16S rRNA gene has enabled a new level of analysis of complex bacterial communities known as microbiomes. If something does not make sense now dont worry. I am a microbial ecologist which means I study how microbes interact with each other and their environment.
Twitter Facebook Google Or copy paste this link into an email or IM. Gut or saliva have proven difficult or even impossible to grow in. We will cover everything shown here in greater detail later.
Previously to study microbes bacteria archaea protists algae fungi and even micro-animals it was necessary to grow them in a laboratory. Ad Learn R by doing 62 interactive coding exercises online. Microbiome data analysis.
An overview of the analysis steps implemented. This vignette provides a brief overview with example data sets from published microbiome profiling studies. R version 410 2021-05-18.
Fork the repository clone it modify the tutorials and make a pull request. Erhalte deinen Bericht und erfahre wie sich die Bakterien auf deine Gesundheit auswirken. This primer provides a concise introduction to conducting applied analyses of microbiome data in R.
A tutorial on how to use Plotlys R graphing library for microbiome data analysis and visualization. Now we have loaded the required files we will be using those OTU. Microbiome Analysis in R.
While this primer does not require extensive knowledge of programming in R the user is expected to install R and all packages required for this primer. If you are a moderator please see our troubleshooting guide. This is a short example analysis to give you a feel for how metacoder and taxa are used in microbiome analysis.
2013 OKeefe et al. We provide examples of using the R packages dada2 phyloseq DESeq2 ggplot2 structSSI and vegan to filter visualize and test microbiome data. However it is still difficult to perform data mining fast and efficiently.
Analysis of beta diversity. This is a tutorial to analyze microbiome data with R. Ad Mittels Next-Generation-DNA-Sequenzierung werden alle Bakterien in deinem Stuhl untersucht.
Key words Graph theory igraph Microbial co-occurrence Microbiome Network OTU table R RStudio. A list of R environment based tools for microbiome data exploration statistical analysis and visualization As a beginner the entire process from sample collection to analysis for sequencing data is a daunting task. A reliable alternative to popular microbiome analysis R packages.
By Nabiilah Ardini Fauziyyah. 3 years ago. The microbiome R packagefacilitates exploration and analysis of microbiome profiling data in particular 16S taxonomic profiling.
In microbial community ecology with the development of high-throughput sequencing techniques the increasing data amount and complexity make the community data analysis and management a challenge. Normalizing count data 4. Hide Comments Share Hide Toolbars Post on.
Leo Lahti Sudarshan Shetty et al. To initiate reproducible documentation do the following in RStudio. We provide standard tools as well as novel extensions on standard analyses to improve interpretability and the analysts ability to communicate results all while maintaining object malleability to encourage open source collaboration.
Introduction to the microbiome R package 1 Introduction The microbiome R package facilitates phyloseq-based exploration and analysis of taxonomic profiling data. A more comprehensive tutorial is available on-line. Here we describe in detail and step by step the process of building analyzing and visualizing microbiome networks from operational taxonomic unit OTU tables in R and RStudio using several different approaches and extensively commented code snippets.
Bacterial composition and abundance were analyzed. Installation Launch RRStudio and install the microbiome R package see installation instructions. Ad Learn R by doing 62 interactive coding exercises online.
I see that the ordination plot of Unweighted unifrac suggests not much difference in microbiome in terms of composition. Downstream plotting and analysis of 16s microbiome data in R using phyloseq and ggplot This beginner-friendly tutorial will allow you to create publication-level graphs and convert phyloseq objects into dataframes for easier manipulation and analysis. In this study the R.
There has been a lot of R packages created for the microbiome profiling analysis. Tutorial 16S microbiome analysis of ATCC R Microbiome Standards ChunLab Inc. But a PERMANOVA of unweighted unifrac says that samples are significantly different.
A more comprehensive tutorial is available. Open a new Rmarkdown html file Convert that html file with the knit button Modify the file and knit again to make your own reproducible report. A more comprehensive tutorial is available on-line.
R6 Class to store and analyze data. 8 min read How to give life to your microbiome data. Erhalte deinen Bericht und erfahre wie sich die Bakterien auf deine Gesundheit auswirken.
Ad Mittels Next-Generation-DNA-Sequenzierung werden alle Bakterien in deinem Stuhl untersucht. Moving microbes from the 50 percent human artscience project. Tutorials for microbiome analysis - GitHub Pages.
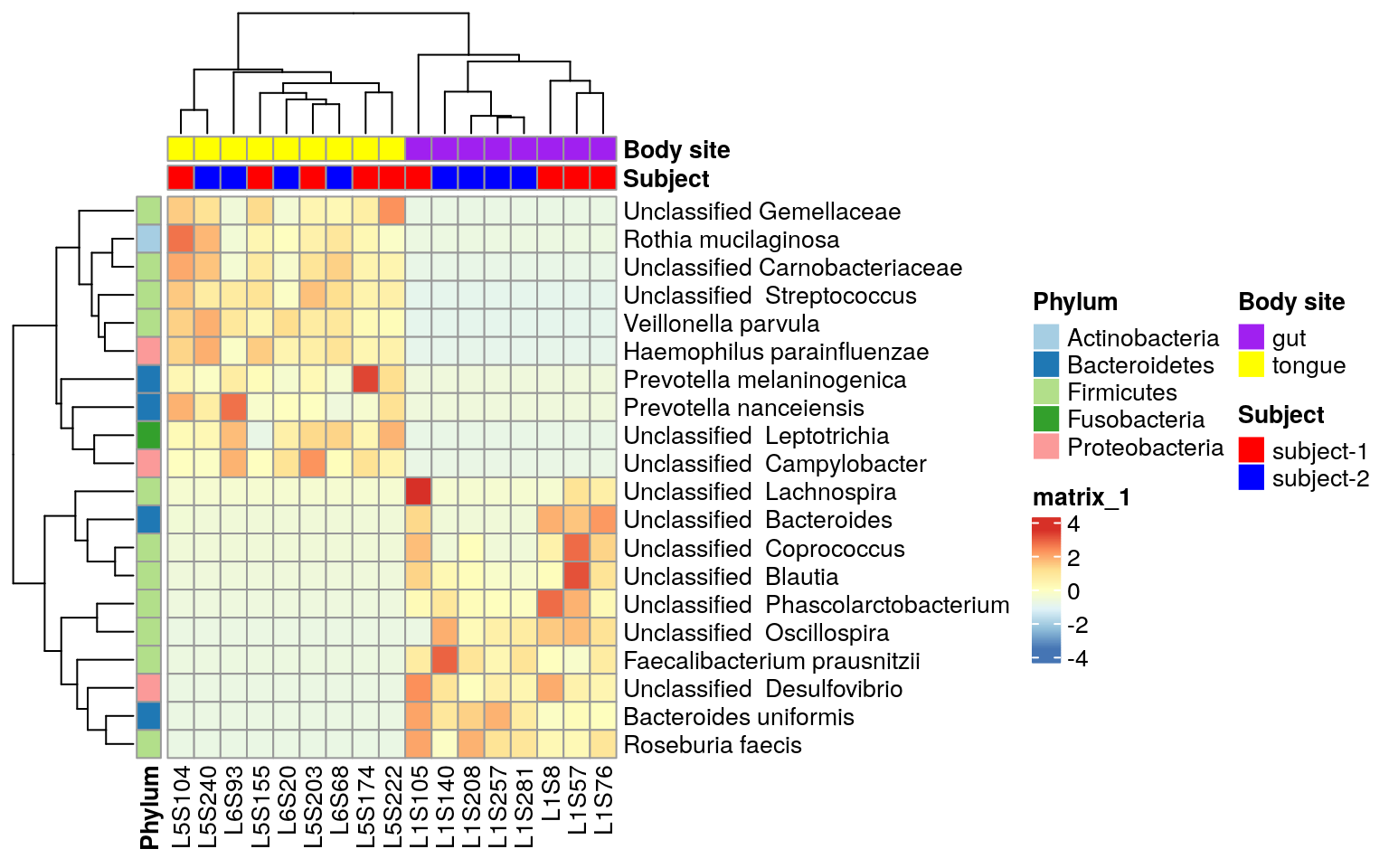
Tutorial For Microbiome Analysis In R Yan Hui
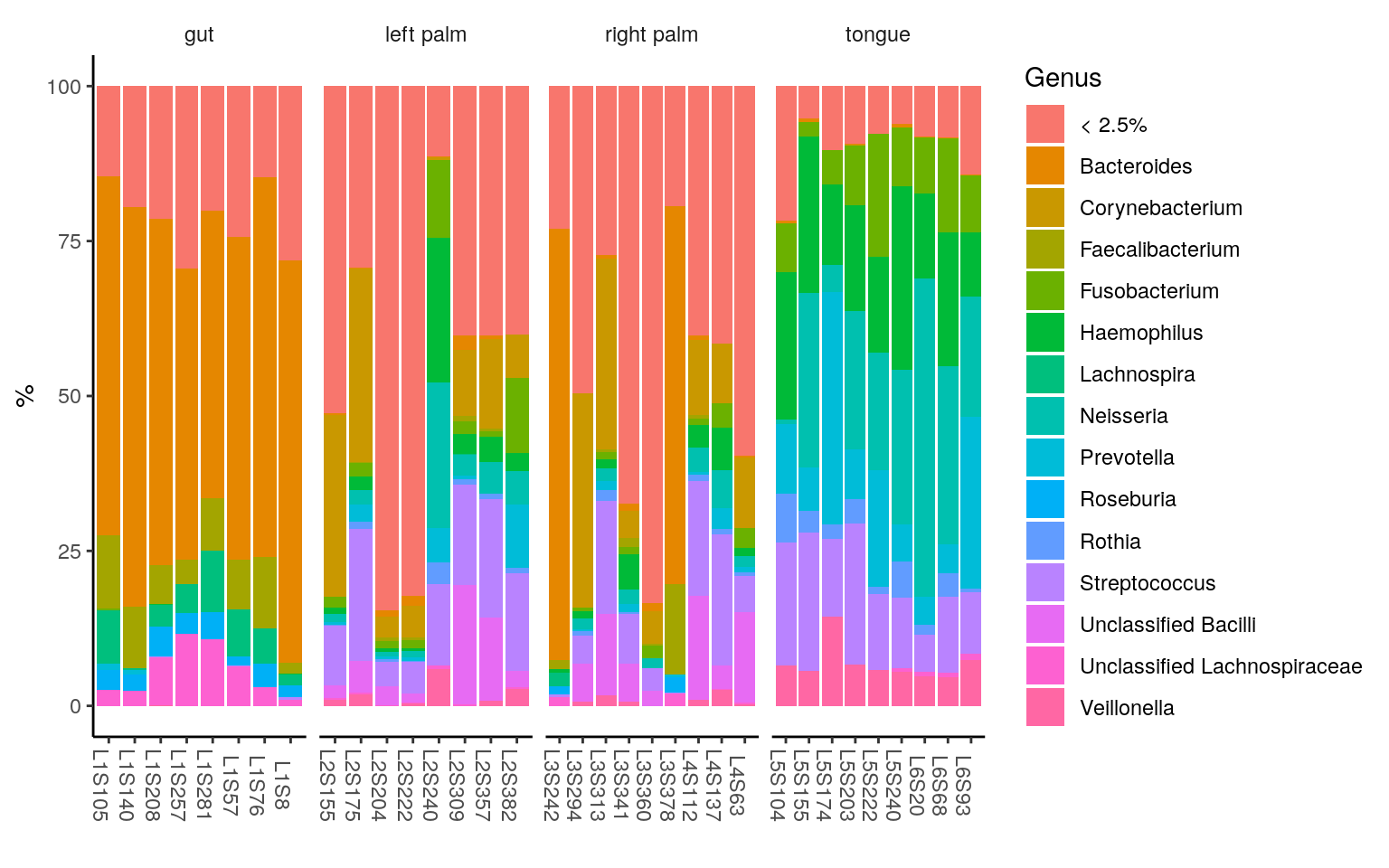
Tutorial For Microbiome Analysis In R Yan Hui
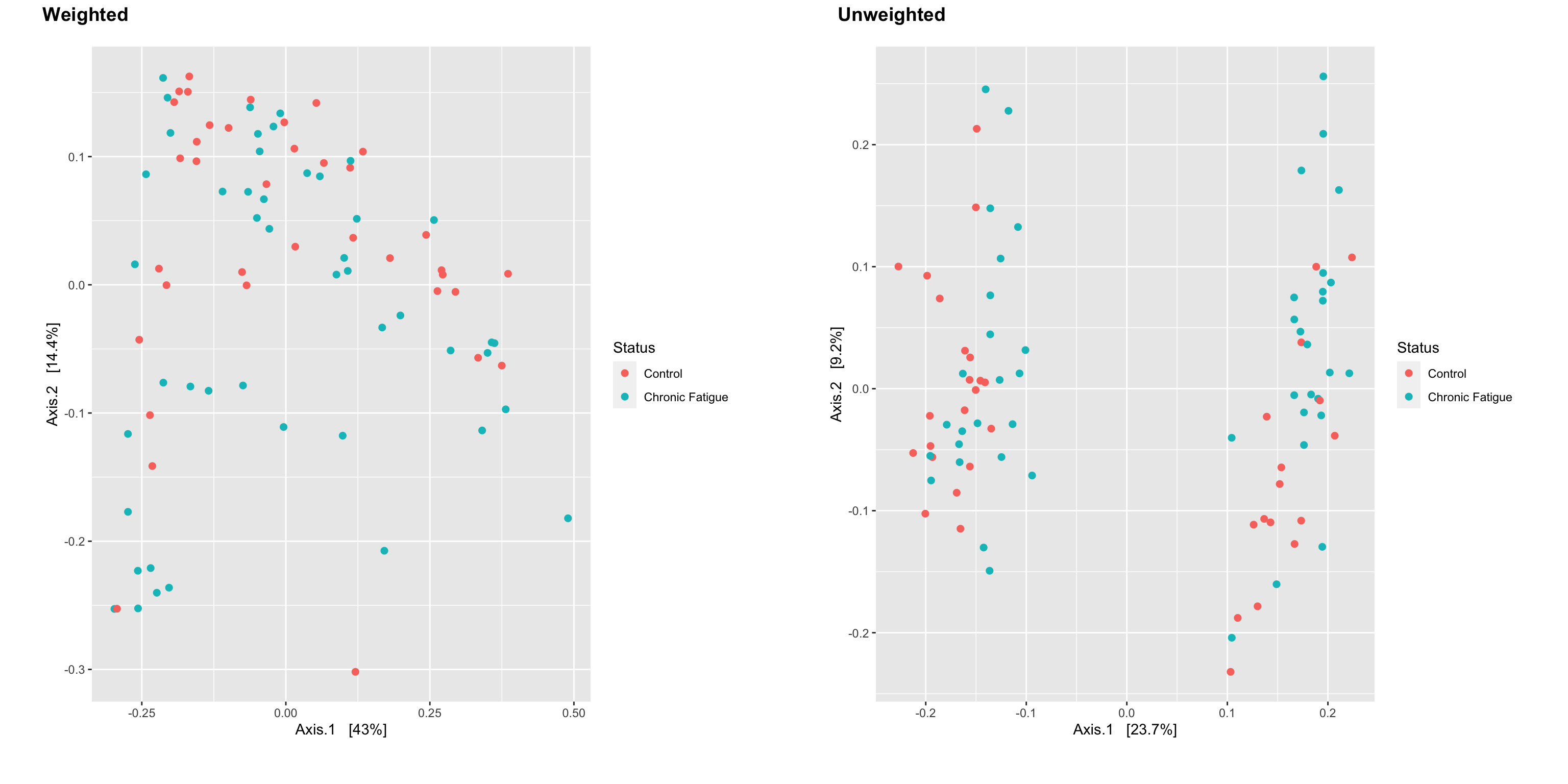
Introduction To The Statistical Analysis Of Microbiome Data In R Academic

Making Heatmaps With R For Microbiome Analysis The Molecular Ecologist
Phyloseq Explore Microbiome Profiles Using R
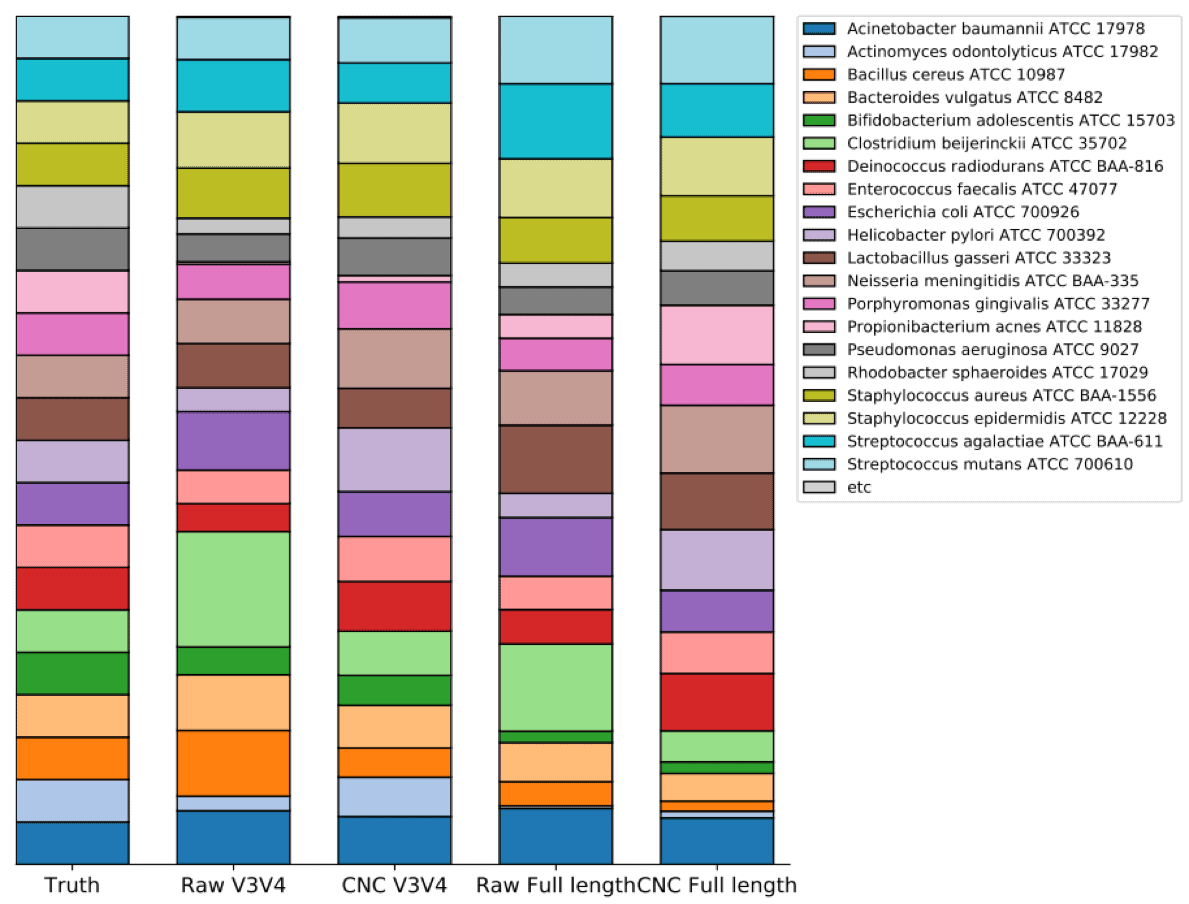
Tutorial 16s Microbiome Analysis Of Atcc R Microbiome Standards Ezbiocloud Help Center
0 comments
Post a Comment